Logistic Regression
- Coding practice
- ~60 Minutes
- Participant survey

PASSENGERS
If you need to load the passengers data set, run this:
source("http://ajstamm.github.io/titanic/workshops/regression/import-passengers.R")
Note: source() runs all code in the file.

Learning Objectives
- Definition
- Changing survived to died
- Logistic Regression
- Visualization
- Odds Ratio
Logistic Regression
- Model with a categorical dependent variable. Today's example is binary: "0" = alive; "1" = dead
- Estimates the probability of a binary response based on predictor variables to determine how much the presence of a risk factor affects the probability of a given outcome.
- For more information, see Wikipedia
Survived
Dependent variable (outcome): what we want to predict.
tbl_surv <- table(passengers$survived)
tbl_surv
0 1
140 110
## table above as percentages
100*prop.table(tbl_surv)
0 1
56 44
Died!
- New column.
- Easier to interpret risk factors.
- Easier to type
diedthansurvived.
passengers$died <- 1
passengers$died[passengers$survived == 1] <- 0
## Check if survived and died are opposites.
## head() shows the first 6 rows of your table
head(passengers[, c("survived", "died")])
survived died
1 0 1
2 1 0
3 1 0
4 1 0
5 0 1
6 0 1
Logistic Regression: Categorical
Independent variable: sex; Dependent variable: died
## Cannot have NAs in our data.
sum(is.na(passengers$sex))
[1] 0
sum(is.na(passengers$died))
[1] 0
## We can only graph numeric variables, not categorical.
passengers$is_male <- 0
passengers$is_male[passengers$sex == "male"] <- 1
logit_sex <- glm(formula=died~is_male, family=binomial,
data=passengers)
To what extent does is_male explain died?
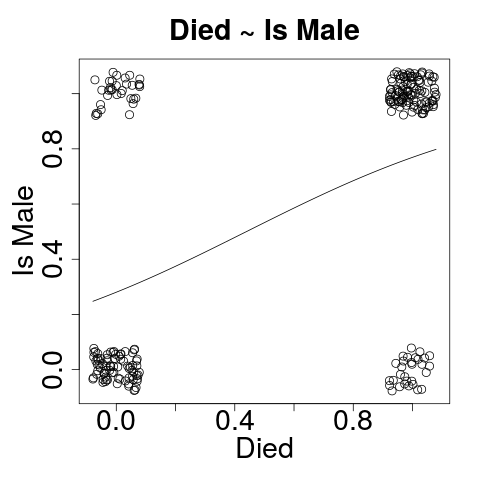
Died~Sex
- Formula: died as a function of sex
- Sex is a statistically significant predictor of died.
- Reduces residual deviance and AIC.
summary(logit_sex)
Call:
glm(formula = died ~ is_male, family = binomial, data = passengers)
Deviance Residuals:
Min 1Q Median 3Q Max
-1.7125 -0.8112 0.7244 0.7244 1.5948
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -0.9426 0.2152 -4.380 1.19e-05 ***
sexmale 2.1466 0.2928 7.332 2.27e-13 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 342.96 on 249 degrees of freedom
Residual deviance: 281.47 on 248 degrees of freedom
AIC: 285.47
Number of Fisher Scoring iterations: 4
Visualizing: Died~Sex
plot(jitter(passengers$died, .4),
jitter(passengers$is_male, .4))
curve(predict(logit_sex, data.frame(is_male=x), type="response"),
add=TRUE)
Logistic Regression: Continuous
Independent variable: age; Dependent variable: died
## Cannot have NAs in our data.
sum(is.na(passengers$age))
[1] 0
sum(is.na(passengers$died))
[1] 0
logit_age <- glm(formula=died~age, family=binomial,
data=passengers)
To what extent does the variable age explain died?
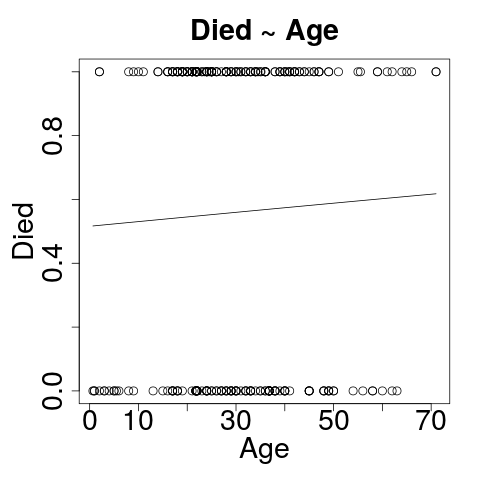
Died ~ Age
- Formula: died as a function of age
- Age is not a statistically significant predictor of died!
AlmostNo difference to residual deviance or AIC.
summary(logit_age)
Call:
glm(formula = died~age, family = binomial, data = passengers)
Deviance Residuals:
Min 1Q Median 3Q Max
-1.366 -1.273 1.037 1.089 1.145
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 0.063888 0.303464 0.211 0.833
age 0.005861 0.009127 0.642 0.521
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 342.96 on 249 degrees of freedom
Residual deviance: 342.55 on 248 degrees of freedom
AIC: 346.55
Number of Fisher Scoring iterations: 4
Visualizing: Died~Age
plot(passengers$age, passengers$died)
curve(predict(g_age, data.frame(age=x), type="response"), add=TRUE)
Yeah - That's supposed to be curved.
Let's Cheat for a moment
- GAM: Generalized Additive Model
- Several confounders here. What can we do?
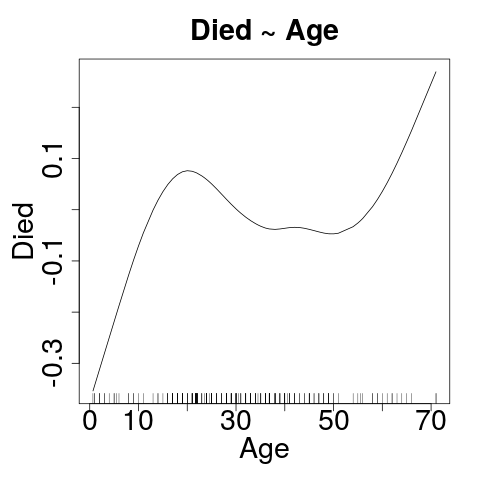
Click on the image to make it easy to really LOOK at it.
Suggestions?
How can we use age to predict died?
New Factors
1. Explore. 2. Learn new tricks.
## You've seen this:
prop.table(table(passengers$died))
0 1
0.44 0.56
## Indexing / Filtering: What percent of passengers
## five years of age or younger died?
prop.table(table(passengers$died[passengers$age <= 5]))
0 1
0.8181818 0.1818182
Note: I'm TIRED of typing passengers.
New Tricks
with |
&, | |
round |
## - Only write passengers once
## - Percent of passengers died older than 5 and younger than 11.
with(passengers,
prop.table(table(died[age > 5 & age <= 10])))
0 1
0.5714286 0.4285714
## Change proportion to percent and round:
with(passengers,
round(prop.table(table(died[age > 5 & age <= 10]))*100))
Age Group, Not Age
## Let's create a new factor:
passengers$age_group <- NA
passengers$age_group[passengers$age <= 10] <- "00-10"
passengers$age_group[passengers$age > 10 &
passengers$age <= 20] <- "11-20"
passengers$age_group[passengers$age > 20 &
passengers$age <= 30] <- "21-30"
passengers$age_group[passengers$age > 30 &
passengers$age <= 40] <- "31-40"
passengers$age_group[passengers$age > 40 &
passengers$age <= 50] <- "41-50"
passengers$age_group[passengers$age > 50 &
passengers$age <= 60] <- "51-60"
passengers$age_group[passengers$age > 60] <- "61+"
Complex Tables
## This should be familiar by now:
with(passengers,
round(prop.table(table(age_group, died),1)*100,1))
## But this is nice!
multi_tbl <- with(passengers,
round(prop.table(100 *
table(age_group, sex, died), 1), 1))
ftable(multi_tbl)
died 0 1
age_group sex
00-10 female 27.8 22.2
male 44.4 5.6
11-20 female 24.3 13.5
male 5.4 56.8
21-30 female 31.3 14.5
male 10.8 43.4
31-40 female 41.7 10.0
male 10.0 38.3
41-50 female 21.2 9.1
male 18.2 51.5
51-60 female 40.0 0.0
male 10.0 50.0
61+ female 11.1 0.0
male 11.1 77.8
 Your Turn!
Your Turn!
Is age_group a statistically significant predictor of died?
No peeking!
Died~Age_Group Is Better
logit_age <- glm(formula=died~age_group, family=binomial,
data=passengers)
summary(logit_age)
Call:
glm(formula = died ~ age_group, family = binomial, data = passengers)
Deviance Residuals:
Min 1Q Median 3Q Max
-1.734 -1.177 0.840 1.047 1.601
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -0.9555 0.5262 -1.816 0.06941 .
age_group11-20 1.8157 0.6374 2.849 0.00439 **
age_group21-30 1.2714 0.5713 2.226 0.02604 *
age_group31-40 0.8888 0.5862 1.516 0.12948
age_group41-50 1.3863 0.6355 2.181 0.02915 *
age_group51-60 0.9555 0.8228 1.161 0.24550
age_group61+ 2.2083 0.9591 2.303 0.02130 *
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 342.96 on 249 degrees of freedom
Residual deviance: 330.08 on 243 degrees of freedom
AIC: 344.08
Number of Fisher Scoring iterations: 4
Tweak these groups to reduce the residuals further.
But don't overfit.
 Your Turn!
Your Turn!
How are these two models different?
logit_complex <- glm(formula =
died ~ age_group+is_male,
family = binomial,
data = passengers)
summary(logit_complex)
logit_complex <- glm(formula =
died ~ age_group*is_male,
family = binomial,
data = passengers)
summary(logit_complex)
?formula
Odds Ratio
- For epidemiologists.
- No Base R odds ratio function.
- Easy to calculate.

Odds Ratio (OR)
Quantifies how strongly the presence or absence of property A is associated with the presence or absence of property B in a given population.
Question: how is being male associated with dying?
Logistic regression calculates OR, not RR.
OR: First Steps
- Want odds of dying for two different groups.
-
Calculate a 2x2 table: died x is_male
This table is backwards from most text books. - Invert the table, or do the math right.
## Remember: Rows (died), Columns (is_male)
t_male <- with(passengers,
table(died, is_male, dnn=c("died","is_male")))
t_male
is_male
died 0 1
0 77 33
1 30 110
## Those numbers alone tell you alot.
Odds of dying
| Man: |
110/33
|
| Woman: |
30/77
|
The ratio of these two odds:
(110/33)/(30/77)
|
or . . . |
(110*77)/(33*30)
|
- The Odds Ratio in our sample is 8.555555555555555 . . .6
- That is an enormous risk factor.
 Your Turn!
Your Turn!
Can you figure out how to do that with R?
It is just some fancy indexing trickery.
OR: Algebraic Method
## Remember:
t_male
is_male
died 0 1
0 77 33
1 30 110
Thus, our solution . . .
## Remember, you can use R like a giant calculator:
(t_male[2,2] * t_male[1,1]) / (t_male[2,1]*t_male[1,2])
[1] 8.555556
## This does not come with confidence intervals.
Pop Quiz!
What is the relationship between OR and logistic regression?
-
The
log()of the OR == the coefficient from the logit model. - Which means . . .
-
The
exp()of the coefficient from the logic model == the OR. - If you can build the logit model, the OR is one step away.
Deriving OR from logit model
## How does this help us?
names(summary(logit_sex))
Deriving OR from logit model
- This is one way to do it.
## Step 1: Access the model coefficients:
summary(logit_sex)$coefficients
Estimate Std. Error z value Pr(>|z|)
(Intercept) -0.942608 0.2152215 -4.379712 1.188363e-05
is_male 2.146581 0.2927698 7.331975 2.267860e-13
## Step 2: Calculate the exponent of the coefficients:
exp( summary(logit_sex)$coefficients )
Estimate Std. Error z value Pr(>|z|)
(Intercept) 0.3896104 1.240137 1.252897e-02 1.000012
is_male 8.5555556 1.340134 1.528397e+03 1.000000
Deriving OR from logit model
- This is another way to do it.
## Step 1: Access the model coefficients:
logit_sex_coef <- summary(logit_sex)$coefficients
## Step 2: Calculate the exponent of the coefficients:
exp( logit_sex_coef )
## - Step 1 returns a vector and saves it as logit_sex_coef.
## - Step 2 calculates the exponent for each item in the vector.
Using R is like building a spaceship with LEGOS!
Assemble little things to build big things.
Next Spring's R Workshops
The following ad is paid for by our sponsors.
- What holds you back from using R?
- Next spring, we plan to offer advanced courses with even more exciting data management tools!
- Stay tuned through EBCoP's blog and your school's announcements.

 Your Turn!
Your Turn!
- Try your new skills in your work.
- At the DOH? Join EBCoP!
- Please complete the workshop survey
- Are there specific skills you want to learn in R? Email us!